Science Beam – using computer vision to extract PDF data
- News
- 3.5K
Blogpost by Daniel Ecer, Data Scientist, and Giuliano Maciocci, Head of Product
There’s a vast trove of science out there locked inside the PDF format. From preprints to peer-reviewed literature and historical research, millions of scientific manuscripts today can only be found in a print-era format that is effectively inaccessible to the web of interconnected online services and APIs that are increasingly becoming the digital scaffold of today’s research infrastructure.
The PDF format is designed for presentation. Extracting key information from PDF files isn’t trivial. We can’t rely on any metadata, paragraphs, or even words since PDF files contain principally four basic components: tokens (which may be characters or words), font glyphs, images, and paths. Higher level elements are inferred from those basic components, as illustrated below.
Daniel Ecer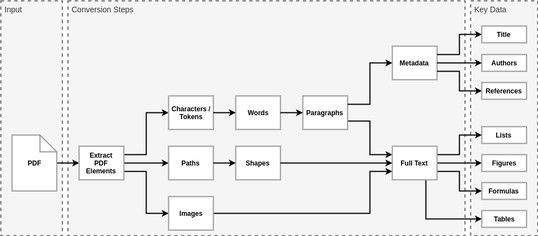
It would therefore certainly be useful to be able to extract all key data from manuscript PDFs and store it in a more accessible, more re-usable format such as XML (of the publishing industry standard JATS variety or otherwise). This would allow for the flexible conversion of the original manuscript into different forms, from mobile-friendly layouts to enhanced views like eLife’s side-by-side view (through eLife Lens). It will also make the research mineable and API-accessible to any number of tools, services, and applications. From advanced search tools to the contextual presentation of semantic tags based on users’ interests, and from cross-domain mash-ups showing correlations between different papers to novel applications like ScienceFair, a move away from PDF and toward a more open and flexible format like XML would unlock a multitude of use cases for the discovery and reuse of existing research.
Services to extract key data from PDFs could also be used in journal submission systems. Such solutions would let authors submit raw PDFs of their work and cut down on the considerable manual labor required to enter the relevant manuscript metadata at submission and turn the whole process from a form-filling exercise to a form-checking one.
Extracting the structure of a research paper, and assigning the correct semantic meaning to it, is only part of the process. To fully and accurately convert a research manuscript PDF to XML, there are many individual processes that must be addressed. Thankfully, there are a number of open-source tools that target those individual processes, including Grobid, CERMINE or ContentMine. These vary in approach from heuristics to machine learning, and thus far none have achieved a level of accuracy and reliability that would solve the problem once and for all.
We are embarking on a project to build on these existing open-source tools and to improve the accuracy of the XML output. One aim of the project is to combine some of the existing tools in a modular PDF-to-XML conversion pipeline that achieves a better overall conversion result compared to using individual tools on their own. In addition, we are experimenting with a different approach to the problem: using computer vision to identify key components of the scientific manuscript in PDF format.
Using computer vision to accurately convert PDF to XML
By first treating the PDF as an image, we’re training a neural network to “see” and recognize the core structure of the PDF, which can then be used to assign the correct metadata to the document’s content.

The type of content in the PDF could be identified by its positioning and formatting
This image is a derivative of and attributed to Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789, and used under Creative Commons Attribution License (CC BY 3.0).
By looking at the “redacted” image here, for example, anyone with a passing familiarity with scientific manuscripts should be able to accurately guess what types of content are in each section. Title, authors, institutions, abstract, and introduction are all indicated simply by their location and shape on the page.
For a computer vision algorithm, this is not such an easy task. In order for it to be able to extract good meta data from the myriad variations in font, layout, and content of PDFs from different sources, we need to train our system with a wide variety of PDFs and their corresponding XML. However, getting hold of raw author-submitted PDFs with sufficiently complete XML is difficult (the XML is usually the result of a lengthy production process at the publisher’s end).
To this end, we will be collaborating with other publishers to collate a broad corpus of valid PDF/XML pairs to help train and test our neural networks. Our hope is that the wide variety of papers and formats in this corpus will help our system learn to deduce the structure of a research paper well enough to be useful in real-world applications.
We invite you to contribute
To help accelerate work in this area, we’re calling on the community to help us move this project forward. We’ve set up a GitHub repository to help structure the project, and we welcome and encourage contributions to any of the key modules identified therein.
The anticipated project outputs can be summarized as:
- Tools and a customizable pipeline for converting PDF to XML
- A trained and tested computer vision model to improve the accuracy of PDF-to-XML conversion
- A training pipeline for the community to run other training data sets for the computer vision model
It will not be possible to publicly release all of the training data, although any public training data, as well as the trained model, can and will be made public (as the model sufficiently abstracts from the original data).
We hope that as a community-driven effort we’ll make more rapid progress towards the vision of transforming PDFs into structured data with high accuracy. Connect with us at innovation@elifesciences.org and continue reading if you’d like to know more about the specifics of our computer vision approach to this task.
Technical overview
Here, we explain the technical background to the computer vision approach of the Science Beam project in more detail and outline the process we are developing.
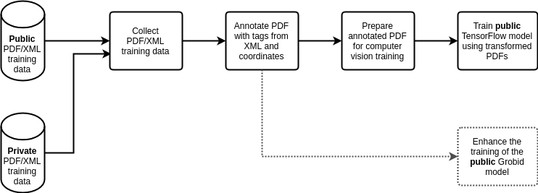
An overview of the model training pipeline, including the training data generation and the training process
Daniel Ecer
So far we have trained the model on 900 PDFs, each from a different journal. These are PDFs from publishers, therefore they have been through production and are more structured already than author versions. We chose this dataset to start with as it is publicly available, including the publisher’s XML, and it is easiest to begin training with well-structured data. Ultimately, for the pipeline to be used for converting author PDFs, such as those shared through preprint servers, into XML for text mining, we will need to continue to train the model on these author-submitted PDFs. These have more variety in their structure, plus the quality and coverage of the accompanying metadata are more variable. First, we need a good model from the best data we have (that from the publisher side).
Step 1: Collect PDF/XML training data
The training data will come as a set of PDFs paired to XML files that describe the metadata for the PDF, including the title, authors, abstracts as well as paragraphs, tables and formulas for the full text. For typeset PDFs, we will have access to JATS XML files that have good coverage of the metadata required (as indicated below), whilst XML accompanying author-submitted PDFs may include less information.

Example JATS XML for a scientific article
This image is a derivative of and attributed to Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789, and used under Creative Commons Attribution License (CC BY 3.0).
Step 2: Annotate PDF with tags from XML and coordinates
The XML files contain the correct tagging for the whole document. For training, we will need to know which PDF elements correspond to which XML tags. In the case of computer vision, we will also need the exact coordinates. Therefore as the first step, we need to use the XML to annotate the PDF elements at the individual character level.
It turns out CERMINE, developed at the Center for Open Science, does exactly that. CERMINE first identifies individual words and text blocks (zones) and then assigns the best matching tag to the whole zone. We aim to improve on that, by adding greater granularity such that text blocks can contain multiple tags: for example, authors and author affiliation can be in the same text block, or the title can appear to be in the same block. Currently, we are using the Smith Waterman algorithm (as does CERMINE) using character-level alignment for shorter texts and word-level alignments for longer text.
The output could look like this, where colors represent individual tags:

A potential output image is an annotated SVG using CSS to attribute non-unique but human-distinguishable colours to identified blocks
This image is a derivative of and attributed to Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789, and used under Creative Commons Attribution License (CC BY 3.0).
Step 3: Prepare annotated PDF for computer vision training
After annotating individual PDF elements with their corresponding tag, we can prepare this data for computer vision training. For this stage, we are more interested in the areas than the individual elements: we plan to convert the annotated PDF elements to colored blocks, each representing a separate tag. The output will be a PNG file. We could map them to unique colors that humans can distinguish, like so:
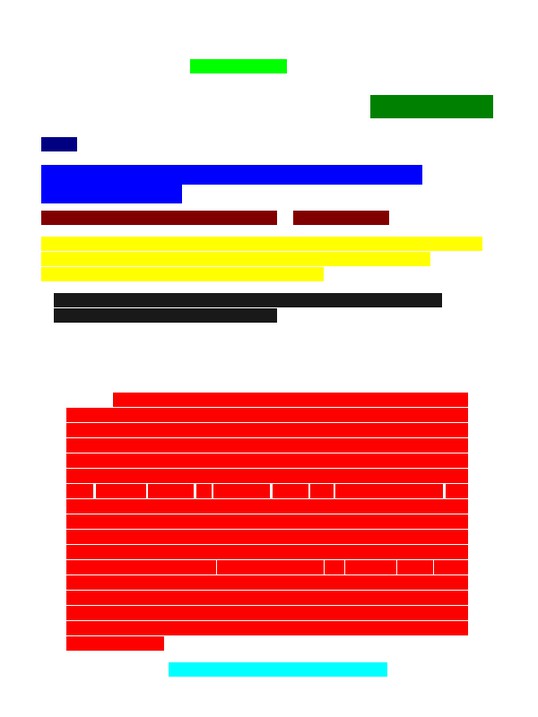
A PNG file with tagged blocks represented by human-distinguishable colors
This image is a derivative of and attributed to Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789, and used under Creative Commons Attribution License (CC BY 3.0).
Step 4: Train public TensorFlow model using transformed PDFs
TensorFlow is an open-source machine learning library that is mostly used to implement deep neural networks. Google provides integration with TensorFlow on its machine learning engine (ML Engine). Using this engine gives us access to Google cloud services that are optimized for training the TensorFlow model and provide access to the Google’s Graphical Processing Units (GPUs), and thus enabling us to train our model quickly whilst minimizing cost.
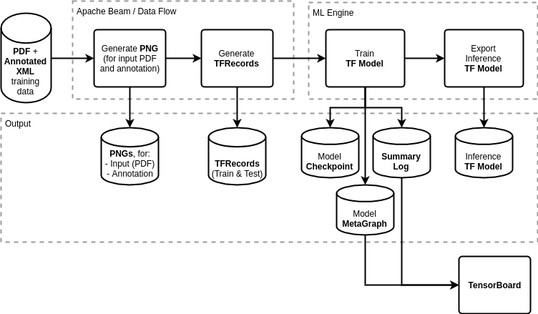
An overview of the public TensorFlow model training pipeline
Daniel Ecer
Before training the TensorFlow model, the training data will be converted into a form that can be more efficiently processed: the annotated PDF training data will be rendered to PNG and then to TFRecords. We plan to execute the preprocessing step via Apache Beam on Google DataFlow. More efficient training will reduce the time it takes to train the model, and thus reduce the cost.
Then the model itself can be trained using Google’s ML Engine, and we could inspect the training progress using TensorBoard.
When assessing the training model progress, we will be looking out for the pixel-based f1 score, a combination of precision and recall, as one reasonable measure of the algorithm’s performance. When we feel the inference model is sufficiently accurate, we will export it for public use.
Implementing the TensorFlow model
How we implement the TensorFlow training model is currently work in progress (see Issue #1 on Github).
In general, this is a semantic segmentation task: the model is meant to detect and annotate regions with tags, such as the title. Provided we give each class a distinct color visible to the human eye, we want to learn the ‘annotated’ image given the input image (rendered PDF). We expect the model to learn that from appearance rather than by being able to read the text. For example:
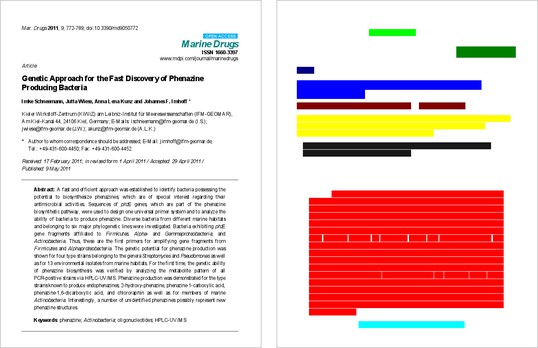
This image is a derivative of and attributed to Schneemann, I.; Wiese, J.; Kunz, A.L.; Imhoff, J.F. Genetic Approach for the Fast Discovery of Phenazine Producing Bacteria. Mar. Drugs 2011, 9, 772-789, and used under Creative Commons Attribution License (CC BY 3.0).
Treating the annotation as a regular RGB image, we already get fairly good results on first samples:
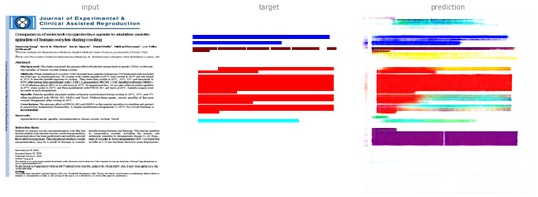
Training computer vision to predict PDF annotation using RGB images
This image is a derivative of and attributed to Yang D, Winslow KL, Nguyen K, Duffy D, Freeman M, Al-Shawaf T. Comparison of selected cryoprotective agents to stabilize meiotic spindles of human oocytes during cooling. Journal of Experimental & Clinical Assisted Reproduction. 2010;7:4, and used under Creative Commons Attribution License (CC BY 2.0).
To find the manuscript title, we would look for the blue colored area.
There are further details about the training so far on the Wiki. We welcome ideas and suggestions from the community as to how to improve this methodology. Please provide feedback on issue #1 of the Github repository.
Enhance the training of the public Grobid
Once the PDF elements are annotated (following step two above), it would become feasible to generate training data for other PDF-to-XML conversion models that use machine learning, including Grobid. Adding our training data into Grobid will improve the accuracy of its algorithm and benefit existing Grobid users. Annotating PDF elements with XML tags (the output data from step 2 above) will help to generate Grobid training data, regardless of the success of our planned TensorFlow model.
Summary
We are beginning work to explore whether computer vision can be used to provide a high-accuracy method to convert PDF to XML. Ideas, feedback, and contributions from the community would be greatly appreciated – we welcome you to explore the project on GitHub (https://github.com/elifesciences/sciencebeam) or you can reach us by email at innovation@elifesciences.org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.


