E-Science Predicts Potential Anti-Coronaviral Drugs and Vaccine Candidates
- News
- 1.7K
The COVID-19 as of today has conquered the whole world. We have witnessed a surge of coronavirus cases with a high death rate. We urgently need effective drugs and vaccines for COVID-19, but what is the quickest way to find it? Currently, the development of a new drug from basic research involves an enormous amount of money and time. Scientists believe that the drug discovery process can speed up significantly using computational biology methods.

This prompted Prof. Rajeev Ahuja, his Ph.D. student, Pritam K. Panda and colleagues at Uppsala University, Sweden in collaboration with SDU, Denmark and KIIT, India to carry out a systematic finding of potent antiviral drugs and vaccine candidates based on state-of-the-art bioinformatics approaches as well as molecular dynamics to determine the binding of different drug molecules.
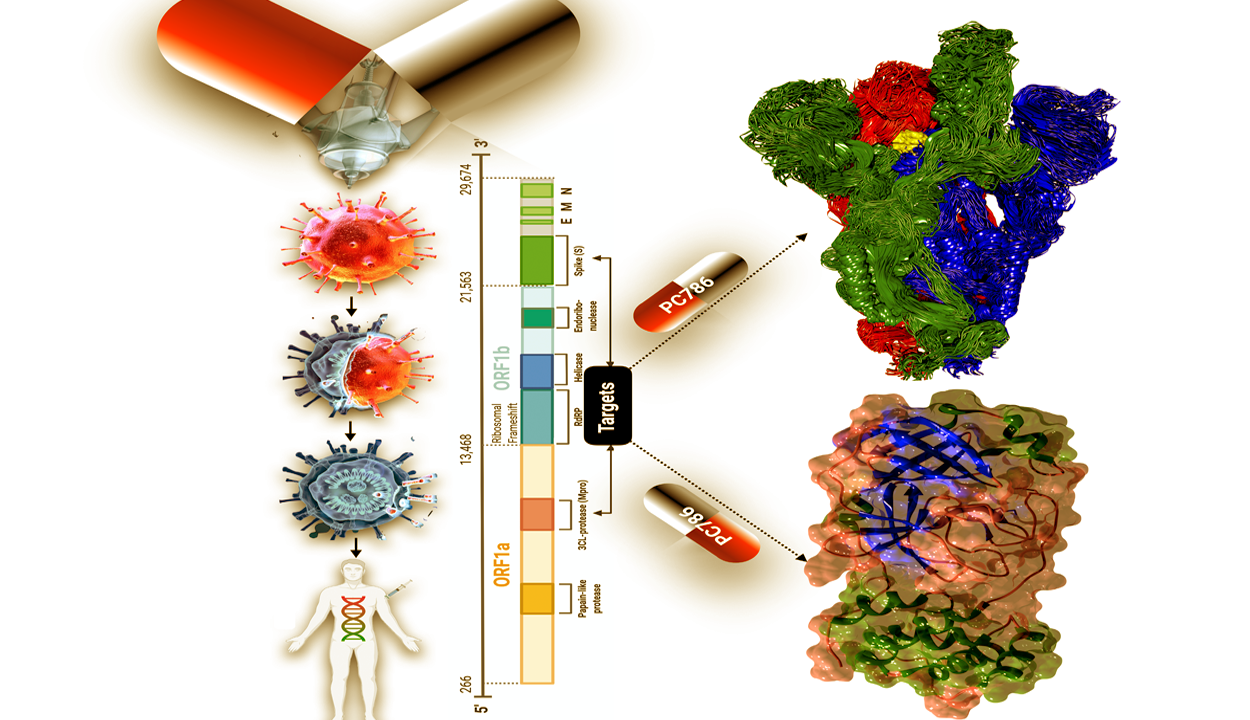
Using bioinformatics approaches, the research team has succeeded in identifying 38 anti-coronaviral drugs from a pool of antiviral drug candidates. This is a relatively novel strategy and virtual screening outfoxes in vivo experiments by many folds in terms of time. They have found that an antiviral inhibitor PC786 targets several SARS-CoV-2 S-protein with high affinity, making it a standout among the antiviral drugs. The findings have been published in the journal ‘Science Advances.’
The study suggests the research team has also identified several additional antiviral drugs with strong binding affinities to the spike protein and main protease, revealing several drugs that may be candidates for further research in efforts to fight SARS-CoV-2. Besides the drug screening approach, they have also proposed potential vaccine candidates (peptides) for SARS-CoV-2 using a structure-based immunoinformatic approach.
The coronavirus needs a special type of protein (the spike protein) to infect a cell. This spike protein acts as a key, to enter into the human cells through an enzyme, ACE2, which is present on the surface of human cells (Present on many issues such as Heart, intestine, and lungs). The virus then fuses with the cell membrane and releases its genetic material into the cell. The spike protein is the sharpest weapon of the virus; its exposed position makes it the preferred point of attack for the immune system.
According to this study, the strategy of a structure-based approach can be employed to screen for drug interactions with certain structural domains and active sites on the virus. This may help scientists identify existing drugs that can be repurposed to treat MERS-CoV, SARS-CoV, Ebola, and HIV. Researchers say, Information about SARS-CoV-2 reported from its recent genome sequencing has revealed key targets for drugs and vaccines, including the spike protein complex, which helps mediate viral entry into host cells, as well as the main protease, an enzyme that enables viral replication and transcription.
The combined drug-designing and immune-informatics approach provide a detailed understanding of the vital structural domains of coronavirus involved in either acting as a drug-binding site or epitope recognition site.
“Our strategy will reduce the translational distance between preclinical test results and clinical outcomes, which is a significant challenge in the rapid development of practical treatment approaches for the on-going coronavirus pandemic,” said Prof. Rajeev Ahuja, corresponding author in this study.
“We do acknowledge several limitations to validate our Insilco proposed work due to the lack of experimental support. PC786 needs clinical validation before putting it to actual use. However, the strategies mentioned above can reduce the resources and expenses for researchers involved directly with the experimental and clinical studies,” he added.
Repurposing of potential drug candidates having a broad-spectrum antiviral activity targeting the viral entry mechanism could be beneficial for clinical use. The current study deals with a similar strategy to find potential drug or vaccine candidates suitable for possible experimental studies targeting the infection pathway of SARS-CoV-2.
“Soon, our long experience in computational science in a combination of Artificial Intelligence (AI) will lead more breakthroughs in data-driven drug discoveries to deal with the current pandemic,” says Prof. Ahuja. The research team included Pritam K. Panda, Murugan N. Arul, Paritosh Patel, Suresh K. Verma, Wei Luo, Horst-Günter Rubahn, Yogendra K. Mishra, Mrutyunjay Suar and Rajeev Ahuja.
If you liked this article, then please subscribe to our YouTube Channel for the latest Science & Tech news. You can also find us on Twitter & Facebook