Toward an “Ultra-Personalized” Therapy for Melanoma
- News
- 1.5K
A new approach to identifying “signposts” on cancer cells may help immune cells eliminate the disease
With new immunotherapy treatments for melanoma, recovery rates have risen dramatically, in some cases to around 50%. But they could be much higher: A new study led by researchers at the Weizmann Institute of Science showed, in lab dishes and animal studies, that a highly personalized approach could help the immune cells improve their abilities to recognize cancer and kill it. The results of this study were published today in Cancer Discovery.
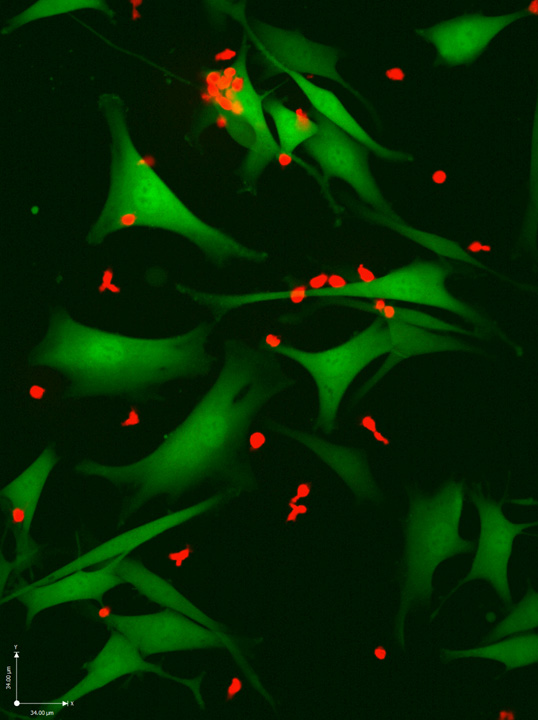
IMAGE: T cells (red) attacking melanoma cells (green). The neo-antigen-specific T cells in this image are especially effective at killing cancer cells Credit: Weizmann Institute of Science
Today’s immunotherapies involve administering antibodies to unlock the natural immune T cells that recognize and kill cancer cells, or else growing and reactivating these T cells outside the body and returning them in a ‘weaponized’ form. “But none of this will kill cancer if the immune cells do not recognize the ‘signposts’ that mark cancer cells as foreign,” says Prof. Yardena Samuels of the Institute’s Molecular Cell Biology Department.
Groups around the world are searching for such signposts – mutated peptides known as neo-antigens on the cancer cells’ outer membranes. Identifying the particular peptides that present themselves to the T cell can then help develop personalized cancer vaccines based on neo-antigen profiles. One of the problems in uncovering neo-antigens in cancers like melanoma, however, is that they are presented by a protein complex called HLA – a complex that can come in thousands of versions, even without the addition of cancerous mutations. Indeed, the algorithms that are often used to search the cancer-cell genome for possible neo-antigens had predicted hundreds of possible candidates.
Samuels, together with her Ph.D. student Shelly Kalaora and in collaboration with Prof Arie Admon of the Technion – Israel Institute of Technology, bypassed the algorithm methods. They used a method Admon had developed to remove the peptides from the melanoma cells’ HLA complex and investigate their interactions with T cells. Says Samuels: “We discovered that tumors present many fewer neo-antigens than we expected.
Our neo-antigen and corresponding T-cell-identification strategies were so robust, our neo-antigen-specific T cells killed 90% of their target melanoma cells both on plates and in mice. This suggests possible clinical applications for the near future. Some of the peptides we identified are neo-antigens that had not even shown up in the algorithm studies; in other words, the method we used, called HLA peptidomics, is really complementary to these methods.
Samuels and Kalaora worked with a team that included Dr. Eytan Ruppin of the National Cancer Institute, USA; Dr. Jennifer Wargo of the University of Texas MD Anderson Cancer Center, Houston, and Prof. Nir Friedman of the Weizmann Institute of Science’s Molecular Cell Biology Department.
For some of the patients, the group had obtained multiple samples, and this enabled them to ask questions about metastasis – for example, whether the same neo-antigens were present on secondary tumors after cancer had spread to other organs. “This is the first time that HLA peptide studies of multiple metastases derived from the same patient have ever been performed.
The significant similarity between the various HLA-peptidomes has strong implications for the process of choosing neo-antigens for patient treatment, showing that it is clearly essential not only to identify the immunogenic peptides presented on the cancer cells but to select the ones that are common to the patient’s metastases. This can then help direct systemic therapeutic responses in patients with multiple metastases,” says Kalaora.
And then the group took their findings one step further: Using natural T cells extracted from 14 of the patients, the researchers identified those that most strongly react with the neo-antigens. They sequenced the genomes of these cells, grew them and tested them in animal models with the patients’ tumor cells. In both lab dish experiments and mice, they showed that the response of the T cells they identified was highly effective in fighting cancer.
Samuels points out that “although this research is experimental right now, the findings are highly relevant to clinical research, as groups around the world have already worked out the basics of developing therapeutic anti-cancer treatments based on neo-antigens. As almost all the neo-antigens detected in patients thus far are individual -and- unique to the particular cancerous tissue — they constitute an ideal class of anti-cancer targets. This would be the ultimate “personalized” cancer therapy—a new drug is created for every patient.”
Also participating in this research were Yochai Wolf, Dr. Itay Tirosh, Nouar Qutob, Polina Greenberg and Ronen Levy of the Molecular Cell Biology Department; Prof. Guy Shakhar, Tali Feferman, Erez Greenstein and Dan Reshef of the Immunology Department; and Ofra Golani of the Life Sciences Core Facilities Department, all of the Weizmann Institute of Science; Eilon Barnea of the Technion – Israel Institute of Technology; Alexandre Reuben, Jianhua Zhang, Xizeng Mao, Xingzhi Song, Chantale Bernatchez, Cara Haymaker, Marie-Andrée Forget, Caitlin Creasy, Brett Carter and Zachary Cooper of the University of Texas MD Anderson Cancer Center; Sushant Patkar of the National Cancer Institute, USA; Juliane Quinkhardt and Tana Omokoko of BioNTech Cell & Gene Therapies, Germany; Prof. Steven Rosenberg of the National Cancer Institute, USA; Michal Lotem of the Hadassah Medical School, Jerusalem, Israel; and Ugur Sahin of University Medical Center of Johannes Gutenberg University, Mainz, Germany.
Prof. Yardena Samuels is the head of the EKARD Institute for Cancer Diagnosis Research; Prof Samuel’s research is supported by the Laboratory in the name of M.E.H Fund established by Margot and Ernst Hamburger; the Weizmann-Brazil Tumor Bank; the European Research Council; the Wagner-Braunsberg Family Melanoma Research Fund; the Comisaroff Family Trust; the Rising Tide Foundation; and Paul and Toni Green. Prof. Samuels is the incumbent of the Knell Family Professorial Chair.
Journal Article
If you liked this article, then please subscribe to our YouTube Channel for the latest Science & Tech news. You can also find us on Twitter & Facebook.


